Dear DEVSIM developer,
Recently, I want to use DEVSIM to simulate the leakage current of a single-side 4H-SiC PN device. But I encounter some problems with the convergence.
This is my simulation process:
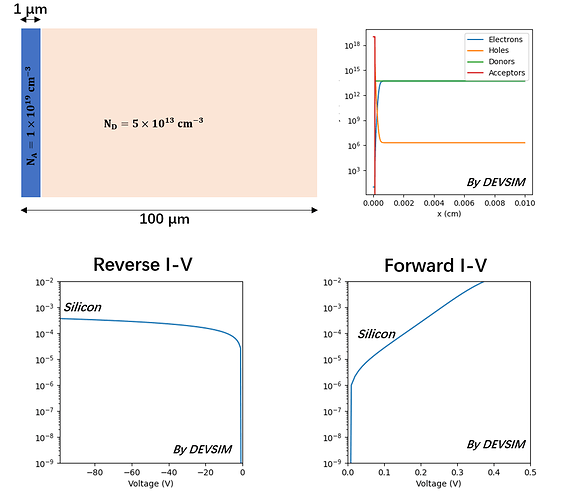
- Firstly, I simulate a single-side Silicon PN device to verify my script and obtain the expected results which are shown below.
- Then, I change the material parameter from silicon to 4H-SiC with the same structure and doping profile. But the simulation is could not be converged. I guess it is caused by the low intrinsic carrier density of 4H-SiC.
Do you have any good suggestions?
The corresponding parameters of 4H-SiC in DEVSIM:
# 4H-SiC parameters used in DEVSIM
eps_sic = 9.76
n_i = 1.18e-8 # #/cm^3
mu_n = 950
mu_p = 125
taun = 2.5e-6
taup = 0.5e-6
My script:
# 4H-SiC single side PN
##############################################################################################################################
# model_create.py
##############################################################################################################################
import devsim
debug = False
def CreateSolution(device, region, name):
'''
Creates solution variables
As well as their entries on each edge
'''
devsim.node_solution(name=name, device=device, region=region)
devsim.edge_from_node_model(node_model=name, device=device, region=region)
def CreateNodeModel(device, region, model, expression):
'''
Creates a node model
'''
result=devsim.node_model(device=device, region=region, name=model, equation=expression)
if debug:
print(("NODEMODEL {d} {r} {m} \"{re}\"".format(d=device, r=region, m=model, re=result)))
def CreateNodeModelDerivative(device, region, model, expression, *vars):
'''
Create a node model derivative
'''
for v in vars:
CreateNodeModel(device, region,
"{m}:{v}".format(m=model, v=v),
"simplify(diff({e},{v}))".format(e=expression, v=v))
def CreateContactNodeModel(device, contact, model, expression):
'''
Creates a contact node model
'''
result=devsim.contact_node_model(device=device, contact=contact, name=model, equation=expression)
if debug:
print(("CONTACTNODEMODEL {d} {c} {m} \"{re}\"".format(d=device, c=contact, m=model, re=result)))
def CreateContactNodeModelDerivative(device, contact, model, expression, variable):
'''
Creates a contact node model derivative
'''
CreateContactNodeModel(device, contact,
"{m}:{v}".format(m=model, v=variable),
"simplify(diff({e}, {v}))".format(e=expression, v=variable))
def CreateEdgeModel (device, region, model, expression):
'''
Creates an edge model
'''
result=devsim.edge_model(device=device, region=region, name=model, equation=expression)
if debug:
print("EDGEMODEL {d} {r} {m} \"{re}\"".format(d=device, r=region, m=model, re=result))
def CreateEdgeModelDerivatives(device, region, model, expression, variable):
'''
Creates edge model derivatives
'''
CreateEdgeModel(device, region,
"{m}:{v}@n0".format(m=model, v=variable),
"simplify(diff({e}, {v}@n0))".format(e=expression, v=variable))
CreateEdgeModel(device, region,
"{m}:{v}@n1".format(m=model, v=variable),
"simplify(diff({e}, {v}@n1))".format(e=expression, v=variable))
def CreateContactEdgeModel(device, contact, model, expression):
'''
Creates a contact edge model
'''
result=devsim.contact_edge_model(device=device, contact=contact, name=model, equation=expression)
if debug:
print(("CONTACTEDGEMODEL {d} {c} {m} \"{re}\"".format(d=device, c=contact, m=model, re=result)))
def CreateContactEdgeModelDerivative(device, contact, model, expression, variable):
'''
Creates contact edge model derivatives with respect to variable on node
'''
CreateContactEdgeModel(device, contact, "{m}:{v}".format(m=model, v=variable), "simplify(diff({e}, {v}))".format(e=expression, v=variable))
def InEdgeModelList(device, region, model):
'''
Checks to see if this edge model is available on device and region
'''
return model in devsim.get_edge_model_list(device=device, region=region)
def InNodeModelList(device, region, model):
'''
Checks to see if this node model is available on device and region
'''
return model in devsim.get_node_model_list(device=device, region=region)
#### Make sure that the model exists, as well as it's node model
def EnsureEdgeFromNodeModelExists(device, region, nodemodel):
'''
Checks if the edge models exists
'''
if not InNodeModelList(device, region, nodemodel):
raise "{} must exist"
emlist = devsim.get_edge_model_list(device=device, region=region)
emtest = ("{0}@n0".format(nodemodel) and "{0}@n1".format(nodemodel))
if not emtest:
if debug:
print("INFO: Creating ${0}@n0 and ${0}@n1".format(nodemodel))
devsim.edge_from_node_model(device=device, region=region, node_model=nodemodel)
##############################################################################################################################
# simple_dd.py
##############################################################################################################################
def CreateBernoulli (device, region):
'''
Creates the Bernoulli function for Scharfetter Gummel
'''
#### test for requisite models here
EnsureEdgeFromNodeModelExists(device, region, "Potential")
vdiffstr="(Potential@n0 - Potential@n1)/V_t"
CreateEdgeModel(device, region, "vdiff", vdiffstr)
CreateEdgeModel(device, region, "vdiff:Potential@n0", "V_t^(-1)")
CreateEdgeModel(device, region, "vdiff:Potential@n1", "-vdiff:Potential@n0")
CreateEdgeModel(device, region, "Bern01", "B(vdiff)")
CreateEdgeModel(device, region, "Bern01:Potential@n0", "dBdx(vdiff) * vdiff:Potential@n0")
CreateEdgeModel(device, region, "Bern01:Potential@n1", "-Bern01:Potential@n0")
def CreateElectronCurrent(device, region, mu_n):
'''
Electron current
'''
EnsureEdgeFromNodeModelExists(device, region, "Potential")
EnsureEdgeFromNodeModelExists(device, region, "Electrons")
EnsureEdgeFromNodeModelExists(device, region, "Holes")
# Make sure the bernoulli functions exist
if not InEdgeModelList(device, region, "Bern01"):
CreateBernoulli(device, region)
Jn = "ElectronCharge*{0}*EdgeInverseLength*V_t*kahan3(Electrons@n1*Bern01, Electrons@n1*vdiff, -Electrons@n0*Bern01)".format(mu_n)
CreateEdgeModel(device, region, "ElectronCurrent", Jn)
for i in ("Electrons", "Potential", "Holes"):
CreateEdgeModelDerivatives(device, region, "ElectronCurrent", Jn, i)
def CreateHoleCurrent(device, region, mu_p):
'''
Hole current
'''
EnsureEdgeFromNodeModelExists(device, region, "Potential")
EnsureEdgeFromNodeModelExists(device, region, "Holes")
# Make sure the bernoulli functions exist
if not InEdgeModelList(device, region, "Bern01"):
CreateBernoulli(device, region)
Jp ="-ElectronCharge*{0}*EdgeInverseLength*V_t*kahan3(Holes@n1*Bern01, -Holes@n0*Bern01, -Holes@n0*vdiff)".format(mu_p)
CreateEdgeModel(device, region, "HoleCurrent", Jp)
for i in ("Holes", "Potential", "Electrons"):
CreateEdgeModelDerivatives(device, region, "HoleCurrent", Jp, i)
##############################################################################################################################
# simple_physics.py
##############################################################################################################################
#TODO: make this a class so that paramters can be changed
contactcharge_node="contactcharge_node"
contactcharge_edge="contactcharge_edge"
ece_name="ElectronContinuityEquation"
hce_name="HoleContinuityEquation"
celec_model = "(1e-10 + 0.5*abs(NetDoping+(NetDoping^2 + 4 * n_i^2)^(0.5)))"
chole_model = "(1e-10 + 0.5*abs(-NetDoping+(NetDoping^2 + 4 * n_i^2)^(0.5)))"
q = 1.6e-19 # coul
k = 1.3806503e-23 # J/K
eps_0 = 8.85e-14 # F/cm^2
#T = 300 # K
eps_sic = 9.76
eps_ox = 3.9
# TODO: make this temperature dependent
n_i = 1.18e-8 # #/cm^3
# constant in our approximation
mu_n = 950
mu_p = 125
def GetContactBiasName(contact):
return "{0}_bias".format(contact)
def GetContactNodeModelName(contact):
return "{0}nodemodel".format(contact)
def PrintCurrents(device, contact):
'''
print out contact currents
'''
# TODO add charge
contact_bias_name = GetContactBiasName(contact)
electron_current= devsim.get_contact_current(device=device, contact=contact, equation=ece_name)
hole_current = devsim.get_contact_current(device=device, contact=contact, equation=hce_name)
total_current = electron_current + hole_current
voltage = devsim.get_parameter(device=device, name=GetContactBiasName(contact))
print("{0}\t{1}\t{2}\t{3}\t{4}".format(contact, voltage, electron_current, hole_current, total_current))
#####
##### Constants
#####
def SetSiliconCarbideParameters(device, region, T):
'''
Sets physical parameters assuming constants
'''
#### TODO: make T a free parameter and T dependent parameters as models
devsim.set_parameter(device=device, region=region, name="Permittivity", value=eps_sic * eps_0)
devsim.set_parameter(device=device, region=region, name="ElectronCharge", value=q)
devsim.set_parameter(device=device, region=region, name="n_i", value=n_i)
devsim.set_parameter(device=device, region=region, name="T", value=T)
devsim.set_parameter(device=device, region=region, name="kT", value=k * T)
devsim.set_parameter(device=device, region=region, name="V_t", value=k*T/q)
devsim.set_parameter(device=device, region=region, name="mu_n", value=mu_n)
devsim.set_parameter(device=device, region=region, name="mu_p", value=mu_p)
#default SRH parameters
devsim.set_parameter(device=device, region=region, name="n1", value=n_i)
devsim.set_parameter(device=device, region=region, name="p1", value=n_i)
devsim.set_parameter(device=device, region=region, name="taun", value=2.5e-6)
devsim.set_parameter(device=device, region=region, name="taup", value=0.5e-6)
def CreateSiliconPotentialOnly(device, region):
'''
Creates the physical models for a Silicon region
'''
if not InNodeModelList(device, region, "Potential"):
print("Creating Node Solution Potential")
CreateSolution(device, region, "Potential")
elec_i = "n_i*exp(Potential/V_t)"
hole_i = "n_i^2/IntrinsicElectrons"
charge_i = "kahan3(IntrinsicHoles, -IntrinsicElectrons, NetDoping)"
pcharge_i = "-ElectronCharge * IntrinsicCharge"
# require NetDoping
for i in (
("IntrinsicElectrons", elec_i),
("IntrinsicHoles", hole_i),
("IntrinsicCharge", charge_i),
("PotentialIntrinsicCharge", pcharge_i)
):
n = i[0]
e = i[1]
CreateNodeModel(device, region, n, e)
CreateNodeModelDerivative(device, region, n, e, "Potential")
### TODO: Edge Average Model
for i in (
("ElectricField", "(Potential@n0-Potential@n1)*EdgeInverseLength"),
("PotentialEdgeFlux", "Permittivity * ElectricField")
):
n = i[0]
e = i[1]
CreateEdgeModel(device, region, n, e)
CreateEdgeModelDerivatives(device, region, n, e, "Potential")
devsim.equation(device=device, region=region, name="PotentialEquation", variable_name="Potential",
node_model="PotentialIntrinsicCharge", edge_model="PotentialEdgeFlux", variable_update="log_damp")
def CreateSiliconPotentialOnlyContact(device, region, contact, is_circuit=False):
'''
Creates the potential equation at the contact
if is_circuit is true, than use node given by GetContactBiasName
'''
# Means of determining contact charge
# Same for all contacts
if not InNodeModelList(device, region, "contactcharge_node"):
CreateNodeModel(device, region, "contactcharge_node", "ElectronCharge*IntrinsicCharge")
#### TODO: This is the same as D-Field
if not InEdgeModelList(device, region, "contactcharge_edge"):
CreateEdgeModel(device, region, "contactcharge_edge", "Permittivity*ElectricField")
CreateEdgeModelDerivatives(device, region, "contactcharge_edge", "Permittivity*ElectricField", "Potential")
contact_model = "Potential -{0} + ifelse(NetDoping > 0, \
-V_t*log({1}/n_i), \
V_t*log({2}/n_i))".format(GetContactBiasName(contact), celec_model, chole_model)
contact_model_name = GetContactNodeModelName(contact)
CreateContactNodeModel(device, contact, contact_model_name, contact_model)
# Simplify it too complicated
CreateContactNodeModel(device, contact, "{0}:{1}".format(contact_model_name,"Potential"), "1")
if is_circuit:
CreateContactNodeModel(device, contact, "{0}:{1}".format(contact_model_name,GetContactBiasName(contact)), "-1")
if is_circuit:
devsim.contact_equation(device=device, contact=contact, name="PotentialEquation",
node_model=contact_model_name, edge_model="",
node_charge_model="contactcharge_node", edge_charge_model="contactcharge_edge",
node_current_model="", edge_current_model="", circuit_node=GetContactBiasName(contact))
else:
devsim.contact_equation(device=device, contact=contact, name="PotentialEquation",
node_model=contact_model_name, edge_model="",
node_charge_model="contactcharge_node", edge_charge_model="contactcharge_edge",
node_current_model="", edge_current_model="")
def CreateSRH(device, region):
USRH="(Electrons*Holes - n_i^2)/(taup*(Electrons + n1) + taun*(Holes + p1))"
Gn = "-ElectronCharge * USRH"
Gp = "+ElectronCharge * USRH"
CreateNodeModel(device, region, "USRH", USRH)
CreateNodeModel(device, region, "ElectronGeneration", Gn)
CreateNodeModel(device, region, "HoleGeneration", Gp)
for i in ("Electrons", "Holes"):
CreateNodeModelDerivative(device, region, "USRH", USRH, i)
CreateNodeModelDerivative(device, region, "ElectronGeneration", Gn, i)
CreateNodeModelDerivative(device, region, "HoleGeneration", Gp, i)
def CreateECE(device, region, mu_n):
CreateElectronCurrent(device, region, mu_n)
NCharge = "-ElectronCharge * Electrons"
CreateNodeModel(device, region, "NCharge", NCharge)
CreateNodeModelDerivative(device, region, "NCharge", NCharge, "Electrons")
devsim.equation(device=device, region=region, name="ElectronContinuityEquation", variable_name="Electrons",
time_node_model = "NCharge",
edge_model="ElectronCurrent", variable_update="positive", node_model="ElectronGeneration")
def CreateHCE(device, region, mu_p):
CreateHoleCurrent(device, region, mu_p)
PCharge = "ElectronCharge * Holes"
CreateNodeModel(device, region, "PCharge", PCharge)
CreateNodeModelDerivative(device, region, "PCharge", PCharge, "Holes")
devsim.equation(device=device, region=region, name="HoleContinuityEquation", variable_name="Holes",
time_node_model = "PCharge",
edge_model="HoleCurrent", variable_update="positive", node_model="HoleGeneration")
def CreatePE(device, region):
pne = "-ElectronCharge*kahan3(Holes, -Electrons, NetDoping)"
CreateNodeModel(device, region, "PotentialNodeCharge", pne)
CreateNodeModelDerivative(device, region, "PotentialNodeCharge", pne, "Electrons")
CreateNodeModelDerivative(device, region, "PotentialNodeCharge", pne, "Holes")
devsim.equation(device=device, region=region, name="PotentialEquation", variable_name="Potential",
node_model="PotentialNodeCharge", edge_model="PotentialEdgeFlux",
time_node_model="", variable_update="log_damp")
def CreateSiliconDriftDiffusion(device, region, mu_n="mu_n", mu_p="mu_p"):
CreatePE(device, region)
CreateBernoulli(device, region)
CreateSRH(device, region)
CreateECE(device, region, mu_n)
CreateHCE(device, region, mu_p)
def CreateSiliconDriftDiffusionAtContact(device, region, contact, is_circuit=False):
'''
Restrict electrons and holes to their equilibrium values
Integrates current into circuit
'''
contact_electrons_model = "Electrons - ifelse(NetDoping > 0, {0}, n_i^2/{1})".format(celec_model, chole_model)
contact_holes_model = "Holes - ifelse(NetDoping < 0, +{1}, +n_i^2/{0})".format(celec_model, chole_model)
contact_electrons_name = "{0}nodeelectrons".format(contact)
contact_holes_name = "{0}nodeholes".format(contact)
CreateContactNodeModel(device, contact, contact_electrons_name, contact_electrons_model)
#TODO: The simplification of the ifelse statement is time consuming
CreateContactNodeModel(device, contact, "{0}:{1}".format(contact_electrons_name, "Electrons"), "1")
CreateContactNodeModel(device, contact, contact_holes_name, contact_holes_model)
CreateContactNodeModel(device, contact, "{0}:{1}".format(contact_holes_name, "Holes"), "1")
#TODO: keyword args
if is_circuit:
devsim.contact_equation(device=device, contact=contact, name="ElectronContinuityEquation",
node_model=contact_electrons_name,
edge_current_model="ElectronCurrent", circuit_node=GetContactBiasName(contact))
devsim.contact_equation(device=device, contact=contact, name="HoleContinuityEquation",
node_model=contact_holes_name,
edge_current_model="HoleCurrent", circuit_node=GetContactBiasName(contact))
else:
devsim.contact_equation(device=device, contact=contact, name="ElectronContinuityEquation",
node_model=contact_electrons_name,
edge_current_model="ElectronCurrent")
devsim.contact_equation(device=device, contact=contact, name="HoleContinuityEquation",
node_model=contact_holes_name,
edge_current_model="HoleCurrent")
##############################################################################################################################
# diode_common.py
##############################################################################################################################
# Make doping a step function
# print dat to text file for viewing in grace
# verify currents analytically
# in dio2 add recombination
#
def CreateMesh(device, region):
'''
Meshing
'''
devsim.create_1d_mesh(mesh="dio")
devsim.add_1d_mesh_line(mesh="dio", pos=0, ps=1e-4, tag="top")
devsim.add_1d_mesh_line(mesh="dio", pos=(1e-4)-(0.5e-4), ps=1e-5, tag="jun_up")
devsim.add_1d_mesh_line(mesh="dio", pos=1e-4, ps=1e-5, tag="mid")
devsim.add_1d_mesh_line(mesh="dio", pos=(1e-4)+(3e-4), ps=1e-5, tag="jun_down")
devsim.add_1d_mesh_line(mesh="dio", pos=100*1e-4, ps=1e-4, tag="bot")
devsim.add_1d_contact (mesh="dio", name="top", tag="top", material="metal")
devsim.add_1d_contact (mesh="dio", name="bot", tag="bot", material="metal")
devsim.add_1d_region (mesh="dio", material="SiC", region=region, tag1="top", tag2="bot")
devsim.finalize_mesh(mesh="dio")
devsim.create_device(mesh="dio", device=device)
def SetParameters(device, region):
'''
Set parameters for 300 K
'''
SetSiliconCarbideParameters(device, region, 300)
def SetNetDoping(device, region):
'''
NetDoping
'''
CreateNodeModel(device, region, "Acceptors", "1.0e19*step(1e-4-x)")
CreateNodeModel(device, region, "Donors", "5.0e13*step(x-1e-4)")
CreateNodeModel(device, region, "NetDoping", "Donors-Acceptors")
def InitialSolution(device, region, circuit_contacts=None):
# Create Potential, Potential@n0, Potential@n1
CreateSolution(device, region, "Potential")
# Create potential only physical models
CreateSiliconPotentialOnly(device, region)
# Set up the contacts applying a bias
for i in devsim.get_contact_list(device=device):
if circuit_contacts and i in circuit_contacts:
CreateSiliconPotentialOnlyContact(device, region, i, True)
else:
###print "FIX THIS"
### it is more correct for the bias to be 0, and it looks like there is side effects
devsim.set_parameter(device=device, name=GetContactBiasName(i), value=0.0)
CreateSiliconPotentialOnlyContact(device, region, i)
def DriftDiffusionInitialSolution(device, region, circuit_contacts=None):
####
#### drift diffusion solution variables
####
CreateSolution(device, region, "Electrons")
CreateSolution(device, region, "Holes")
####
#### create initial guess from dc only solution
####
devsim.set_node_values(device=device, region=region, name="Electrons", init_from="IntrinsicElectrons")
devsim.set_node_values(device=device, region=region, name="Holes", init_from="IntrinsicHoles")
###
### Set up equations
###
CreateSiliconDriftDiffusion(device, region)
for i in devsim.get_contact_list(device=device):
if circuit_contacts and i in circuit_contacts:
CreateSiliconDriftDiffusionAtContact(device, region, i, True)
else:
CreateSiliconDriftDiffusionAtContact(device, region, i)
##############################################################################################################################
# diode_1d.py
##############################################################################################################################
device="MyDevice"
region="MyRegion"
CreateMesh(device=device, region=region)
SetParameters(device=device, region=region)
devsim.set_parameter(device=device, region=region, name="taun", value=2.5e-6)
devsim.set_parameter(device=device, region=region, name="taup", value=0.5e-6)
devsim.set_parameter(name = "extended_solver", value=True)
devsim.set_parameter(name = "extended_model", value=True)
devsim.set_parameter(name = "extended_equation", value=True)
SetNetDoping(device=device, region=region)
devsim.print_node_values(device=device, region=region, name="NetDoping")
InitialSolution(device, region)
# Initial DC solution
devsim.solve(type="dc", absolute_error=1.0, relative_error=1e-10, maximum_iterations=30)
DriftDiffusionInitialSolution(device, region)
###
### Drift diffusion simulation at equilibrium
###
devsim.solve(type="dc", absolute_error=1e10, relative_error=1e-10, maximum_iterations=30)
import matplotlib
import matplotlib.pyplot
fig1=matplotlib.pyplot.figure(num=1,figsize=(4,4))
x=devsim.get_node_model_values(device=device, region=region, name="x")
fields = ("Electrons", "Holes", "Donors", "Acceptors")
for i in fields:
y=devsim.get_node_model_values(device=device, region=region, name=i)
matplotlib.pyplot.semilogy(x, y)
matplotlib.pyplot.xlabel('x (cm)')
matplotlib.pyplot.ylabel('Density (#/cm^3)')
matplotlib.pyplot.legend(fields)
matplotlib.pyplot.savefig("diode_1d_SiC_density.png")
####
#### Ramp the bias to forward
####
forward_v = 0.0
forward_voltage = []
forward_top_current = []
forward_bot_current = []
forward_voltage.append(0.)
forward_top_current.append(0.)
while forward_v < 0.51:
devsim.set_parameter(device=device, name=GetContactBiasName("top"), value=forward_v)
devsim.solve(type="dc", absolute_error=1e10, relative_error=1e-10, maximum_iterations=30)
PrintCurrents(device, "top")
PrintCurrents(device, "bot")
forward_top_electron_current= devsim.get_contact_current(device=device, contact="top", equation="ElectronContinuityEquation")
forward_top_hole_current = devsim.get_contact_current(device=device, contact="top", equation="HoleContinuityEquation")
forward_top_total_current = forward_top_electron_current + forward_top_hole_current
forward_voltage.append(forward_v)
forward_top_current.append(abs(forward_top_total_current))
forward_v += 0.01
print(forward_voltage)
print(forward_top_current)
fig2=matplotlib.pyplot.figure(num=2,figsize=(4,4))
matplotlib.pyplot.semilogy(forward_voltage, forward_top_current)
matplotlib.pyplot.xlabel('Voltage (V)')
matplotlib.pyplot.ylabel('Current (A)')
matplotlib.pyplot.axis([min(forward_voltage), max(forward_voltage), 1e-9, 1e-2])
matplotlib.pyplot.savefig("diode_1d_SiC_IV_forward.png")
####
#### Ramp the bias to Reverse
####
reverse_v = 0.0
reverse_voltage = []
reverse_top_current = []
reverse_bot_current = []
reverse_voltage.append(0.)
reverse_top_current.append(0.)
while reverse_v < 100.0:
devsim.set_parameter(device=device, name=GetContactBiasName("top"), value=0-reverse_v)
devsim.solve(type="dc", absolute_error=1e10, relative_error=1e-10, maximum_iterations=30)
PrintCurrents(device, "top")
PrintCurrents(device, "bot")
reverse_top_electron_current= devsim.get_contact_current(device=device, contact="top", equation="ElectronContinuityEquation")
reverse_top_hole_current = devsim.get_contact_current(device=device, contact="top", equation="HoleContinuityEquation")
reverse_top_total_current = reverse_top_electron_current + reverse_top_hole_current
reverse_voltage.append(0-reverse_v)
reverse_top_current.append(abs(reverse_top_total_current))
reverse_v += 1.0
print(reverse_voltage)
print(reverse_top_current)
fig3=matplotlib.pyplot.figure(num=3,figsize=(4,4))
matplotlib.pyplot.semilogy(reverse_voltage, reverse_top_current)
matplotlib.pyplot.xlabel('Voltage (V)')
matplotlib.pyplot.ylabel('Current (A)')
matplotlib.pyplot.axis([min(reverse_voltage), max(reverse_voltage), 1e-9, 1e-2])
matplotlib.pyplot.savefig("diode_1d_SiC_IV_reverse.png")