Dear Juan,
thank you for your replay and I appreciate your time.
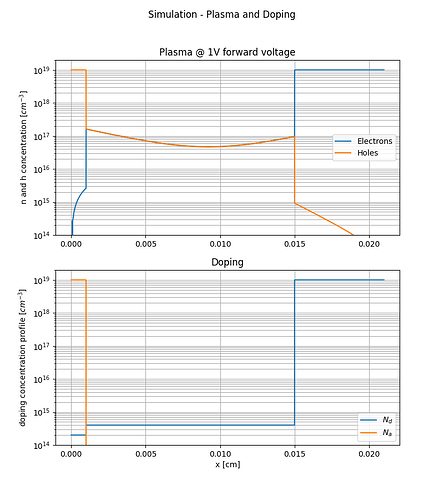
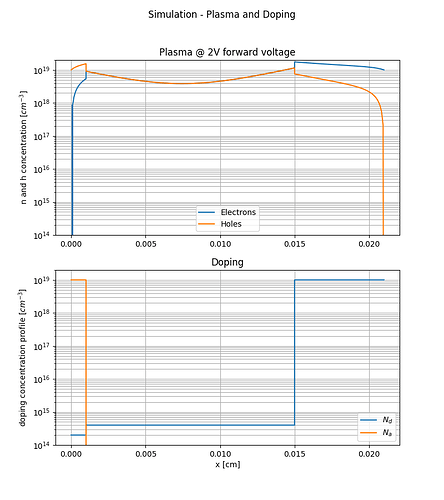
I build a Si pin diode model which works very well when the diode is forward biased as can be seen from the attached figures.
However, when the diode is reversed biased I get the following error at -10V:
error: DEVSIM FATAL: while evaluating node model IntrinsicHoles:Potential on Device: Si_PiN on Region: Region
There was a Overflow floating point exception while evaluating pow(IntrinsicElectrons,(-2))
while evaluating model IntrinsicHoles:Potential: expression (-pow(IntrinsicElectrons,(-2)) * pow(n_i,2) * IntrinsicElectrons:Potential) evaluates to invalid
I did not implement the impact ionization following the example you kindly sent and I don’t think that this error is related to impact ionization because the reverse bias voltage is relativity low (-10V). The introduced pin diode model is supposed to block more that 1200V. I am trying to plot the electric field and potential in the n- drift layer. By the why, the error happens only when trying to use write_devices(file="pin_si_diode_rb", type="vtk"). I am not sure if this information is helpful.
For completeness I am attaching the simulation script. I hope you can advice on what I am missing here.
My best,
Said
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Wed Sep 18 21:41:49 2024
@author: seb
"""
from devsim import *
from devsim.python_packages.simple_physics import *
from devsim.python_packages.ramp import *
from devsim.python_packages.model_create import CreateSolution
import numpy as np
import matplotlib.pyplot as plt
reset_devsim()
contactcharge_edge = "contactcharge_edge"
ece_name = "ElectronContinuityEquation"
hce_name = "HoleContinuityEquation"
# celec_model = "(0.5*abs(NetDoping+(NetDoping^2 + 4 * n_i^2)^(0.5)))"
# chole_model = "(0.5*abs(-NetDoping+(NetDoping^2 + 4 * n_i^2)^(0.5)))"
celec_model = "(1e-9 + 0.5*abs(NetDoping+(NetDoping^2 + 4 * n_i^2)^(0.5)))"
chole_model = "(1e-9 + 0.5*abs(-NetDoping+(NetDoping^2 + 4 * n_i^2)^(0.5)))"
q = 1.6e-19 # coul
k = 1.3806503e-23 # J/K
eps_0 = 8.85e-14 # F/cm^2
eps_si = 11.1
eps_ox = 3.9
n_i = 1.0e10 # #/cm^3
mu_n = 400 #1350 #400
mu_p = 200 #480 #200
tau_n = 2e-6
tau_p = tau_n/3
# Doping
Na_body = 1e+19
Nd_drift= 2e+14
Nd_subs = 1e+19
W_d = 140e-6
name="Si_PiN_Diode"
#All values in cm
def Create2DMesh(device, region):
create_2d_mesh(mesh="dio")
add_2d_mesh_line(mesh="dio", dir="x", pos=0, ps=1e-4) # P_bode
add_2d_mesh_line(mesh="dio", dir="x", pos=10e-4, ps=0.8e-6) # P body
add_2d_mesh_line(mesh="dio", dir="x", pos=80e-4, ps=1e-4) # N_minus
add_2d_mesh_line(mesh="dio", dir="x", pos=150e-4, ps=0.8e-6) # N_minus
add_2d_mesh_line(mesh="dio", dir="x", pos=210e-4, ps=1e-4) # N_plus
add_2d_mesh_line(mesh="dio", dir="y", pos=0, ps=0.5e-4) # y
add_2d_mesh_line(mesh="dio", dir="y", pos=30e-4, ps=0.5e-4) # y
add_2d_mesh_line(mesh="dio", dir="x", pos=-1e-4, ps=1e-4) # top metal
add_2d_mesh_line(mesh="dio", dir="x", pos= 211e-4,ps=1e-4) # bot metal
add_2d_region(mesh="dio", material="Si", region=region)
add_2d_region(mesh="dio", material="SiC", region="air1", xl=-1e-4, xh=0)
add_2d_region(mesh="dio", material="SiC", region="air2", xl=210e-4, xh=211e-4)
add_2d_contact(mesh="dio", name="top", material="metal", region=region, yl=0, yh=30e-4, xl=-1e-4, xh=0, bloat=1e-10)
add_2d_contact(mesh="dio", name="bot", material="metal", region=region, xl=210e-4, xh=211e-4, bloat=1e-10)
finalize_mesh(mesh="dio")
create_device(mesh="dio", device=device)
write_devices(file="si_pin_diode_mesh", type="vtk")
def SetNetDoping(device, region):
'''
NetDoping
'''
CreateNodeModel(device, region, "Acceptors", str(Na_body)+"*step(10e-4-x)") # P doping
CreateNodeModel(device, region, "Donors", str(Nd_drift) + "*(step(-10e-4+x)+step(150e-4-x))" + "+" + str(Nd_subs) + "*step(-150e-4+x)") # N- doping
CreateNodeModel(device, region, "NetDoping", "Donors-Acceptors")
def InitialSolution(device, region, circuit_contacts=None):
# Create Potential, Potential@n0, Potential@n1
CreateSolution(device, region, "Potential")
# Create potential only physical models
CreateSiliconPotentialOnly(device, region)
# Set up the contacts applying a bias
for i in get_contact_list(device=device):
if circuit_contacts and i in circuit_contacts:
CreateSiliconPotentialOnlyContact(device, region, i, True)
else:
###print "FIX THIS"
### it is more correct for the bias to be 0, and it looks like there is side effects
set_parameter(device=device, name=GetContactBiasName(i), value=0.0)
CreateSiliconPotentialOnlyContact(device, region, i)
def DriftDiffusionInitialSolution(device, region, circuit_contacts=None):
####
#### drift diffusion solution variables
####
CreateSolution(device, region, "Electrons")
CreateSolution(device, region, "Holes")
####
#### create initial guess from dc only solution
####
set_node_values(device=device, region=region, name="Electrons", init_from="IntrinsicElectrons")
set_node_values(device=device, region=region, name="Holes", init_from="IntrinsicHoles")
###
### Set up equations
###
CreateSiliconDriftDiffusion(device, region)
for i in get_contact_list(device=device):
if circuit_contacts and i in circuit_contacts:
CreateSiliconDriftDiffusionAtContact(device, region, i, True)
else:
CreateSiliconDriftDiffusionAtContact(device, region, i)
def SetSiliconParameters_my(device, region, T):
"""
Sets physical parameters assuming constants
"""
#### TODO: make T a free parameter and T dependent parameters as models
set_parameter(
device=device, region=region, name="Permittivity", value=eps_si * eps_0
)
set_parameter(device=device, region=region, name="ElectronCharge", value=q)
set_parameter(device=device, region=region, name="n_i", value=n_i)
set_parameter(device=device, region=region, name="T", value=T)
set_parameter(device=device, region=region, name="kT", value=k * T)
set_parameter(device=device, region=region, name="V_t", value=k * T / q)
set_parameter(device=device, region=region, name="mu_n", value=mu_n)
set_parameter(device=device, region=region, name="mu_p", value=mu_p)
# default SRH parameters
set_parameter(device=device, region=region, name="n1", value=n_i)
set_parameter(device=device, region=region, name="p1", value=n_i)
set_parameter(device=device, region=region, name="taun", value=tau_n) #0.2e-6
set_parameter(device=device, region=region, name="taup", value=tau_p) #0.045e-6
set_parameter(device=device, region=region, name="bot", value=0.0) #0.045e-6
if __name__=='__main__':
dev='Si_PiN'
region='Region'
Create2DMesh(dev,region)
SetSiliconParameters_my(dev, region, 300)
# set_parameter(name = "extended_solver", value=True)
# set_parameter(name = "extended_model", value=True)
# set_parameter(name = "extended_equation", value=True)
SetNetDoping(dev, region=region)
InitialSolution(dev, region)
solve(type="dc", absolute_error=1e5, relative_error=1e-12, maximum_iterations=50)
write_devices(file="si_diode_e", type="vtk")
DriftDiffusionInitialSolution(dev, region)
solve(type="dc", absolute_error=1e5, relative_error=1e-12, maximum_iterations=100)
FWB=False # False for reverse bias
fw_voltage=1
bw_voltage=-10
if FWB==True:
rampbias(dev, "top",fw_voltage,0.2,0.001, 60, 1e-12, 1e5, printAllCurrents)
write_devices(file="pin_si_diode_fw", type="vtk")
else:
rampbias(dev, "top",bw_voltage,2,0.01, 60, 1e-12, 1e5, printAllCurrents)
# delete_node_model(device=dev, region=region, name="IntrinsicElectrons")
# delete_node_model(device=dev, region=region, name="IntrinsicHoles")
# for i in np.linspace(0, 10, 20):
# print(f'############### {-i:2.2f} ################')
# set_parameter(device=dev, name=GetContactBiasName("top"), value=-i)
# solve(type="dc", absolute_error=1e5, relative_error=1e-9, maximum_iterations=100)
print('############### finished ################')
write_devices(file="pin_si_diode_rb", type="vtk")
#%%
fig,ax=plt.subplots(2,1,figsize=(8,9))
x=get_node_model_values(device=dev, region=region, name="x")
if FWB==True:
fig.suptitle('Simulation - Plasma and Doping')
y_e=get_node_model_values(device=dev, region=region, name="Electrons")
y_h=get_node_model_values(device=dev, region=region, name="Holes")
y_d=get_node_model_values(device=dev, region=region, name="Donors")
y_a=get_node_model_values(device=dev, region=region, name="Acceptors")
ax[0].set_title(f'Plasma @ {fw_voltage}V forward voltage')
ax[0].semilogy(x,y_e,label='Electrons')
ax[0].semilogy(x,y_h,label='Holes')
ax[0].set_ylim([1e14,2e19])
ax[0].set(ylabel='n and h concentration [$cm^{-3}$]')
ax[0].legend()
ax[0].grid(which='both')
ax[1].set_title('Doping')
ax[1].semilogy(x,y_d,label='$N_d$')
ax[1].semilogy(x,y_a,label='$N_a$')
ax[1].set_ylim([1e14,2e19])
ax[1].set(ylabel='doping concentration profile [$cm^{-3}$]')
ax[1].set(xlabel='x [cm]')
ax[1].legend()
ax[1].grid(which='both')
fig.savefig(f'{name}_at{fw_voltage}V.png')
else:
fig.suptitle('Simulation - Plasma and Doping')
# y_E=get_node_model_values(device=dev, region=region, name="ElectricField")
y_phi=get_node_model_values(device=dev, region=region, name="Potential")
ax[0].set_title('Plasma')
# ax[0].plot(x,y_E,label='E-Field')
ax[0].plot(x,y_phi,label='Potentia.')
# ax[0].set_ylim([1e14,2e19])
ax[0].set(ylabel='n and h concentration [$cm^{-3}$]')
ax[0].legend()
ax[0].grid(which='both')
# ax[1].set_title('Doping')
# ax[1].semilogy(x,y_d,label='$N_d$')
# ax[1].semilogy(x,y_a,label='$N_a$')
# ax[1].set_ylim([1e14,2e19])
# ax[1].set(ylabel='doping concentration profile [$cm^{-3}$]')
# ax[1].set(xlabel='x [cm]')
# ax[1].legend()
# ax[1].grid(which='both')