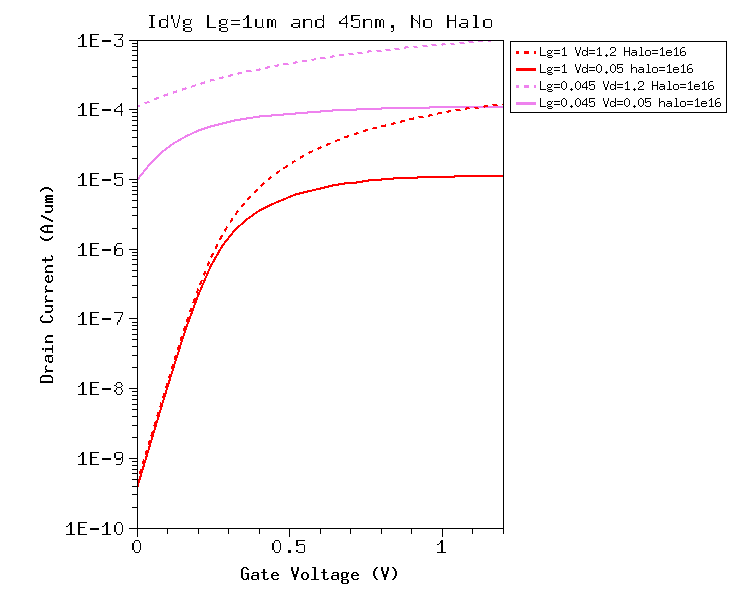
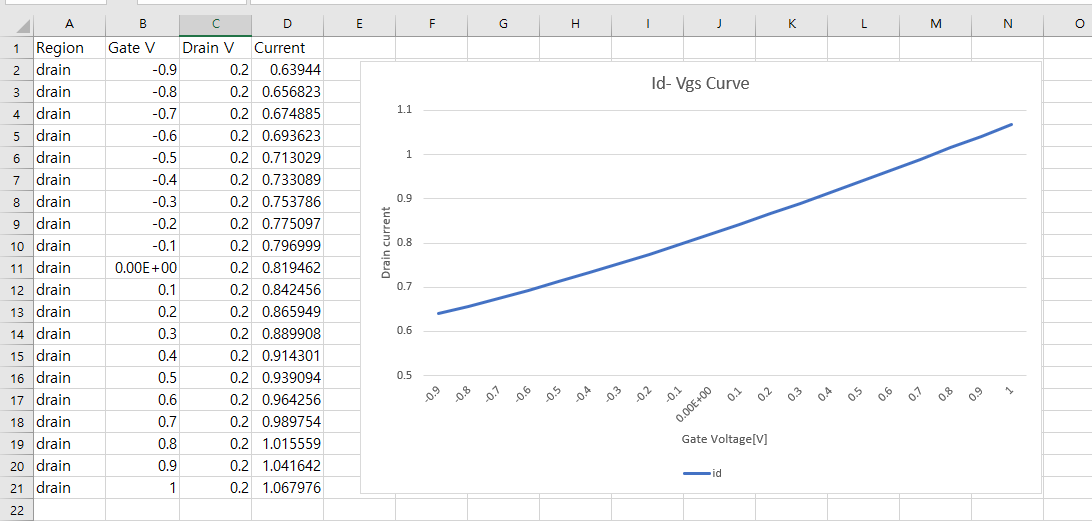
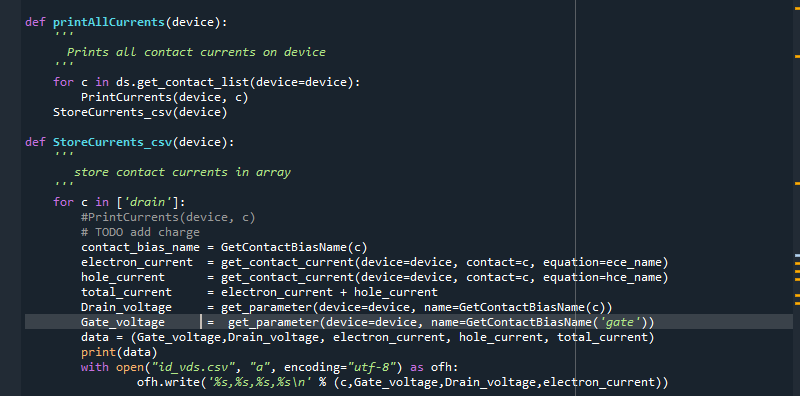
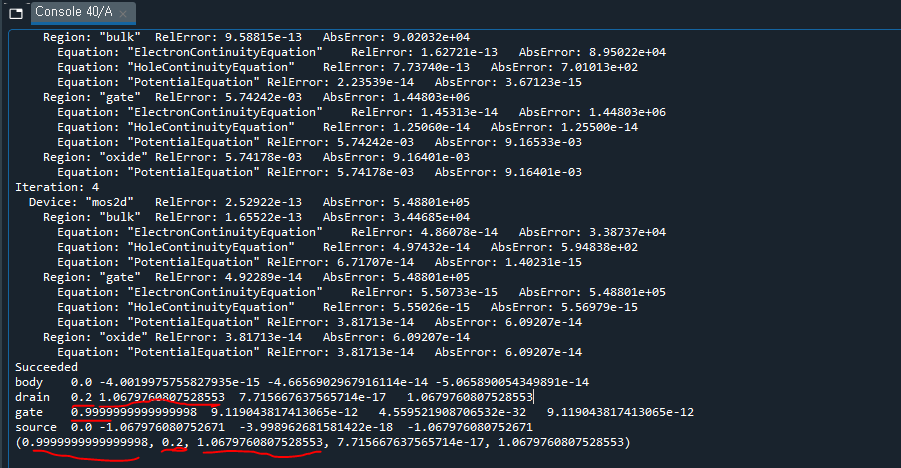
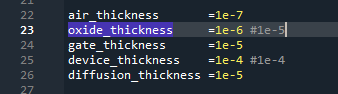
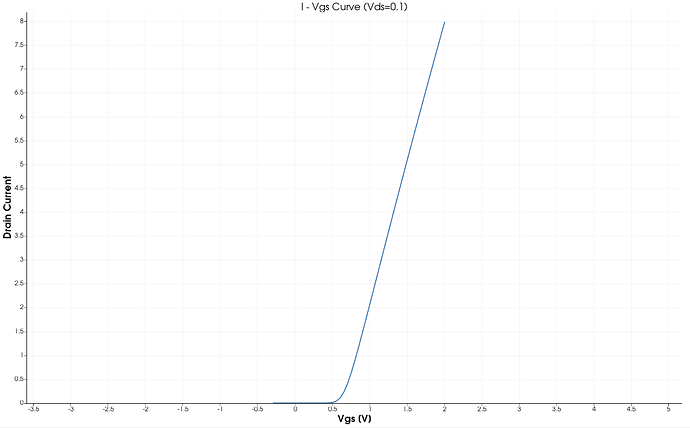
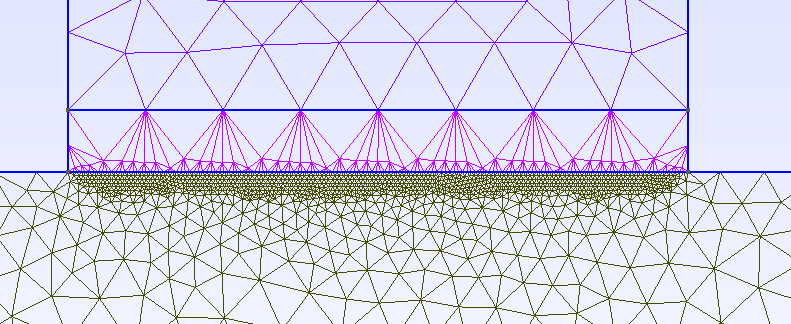
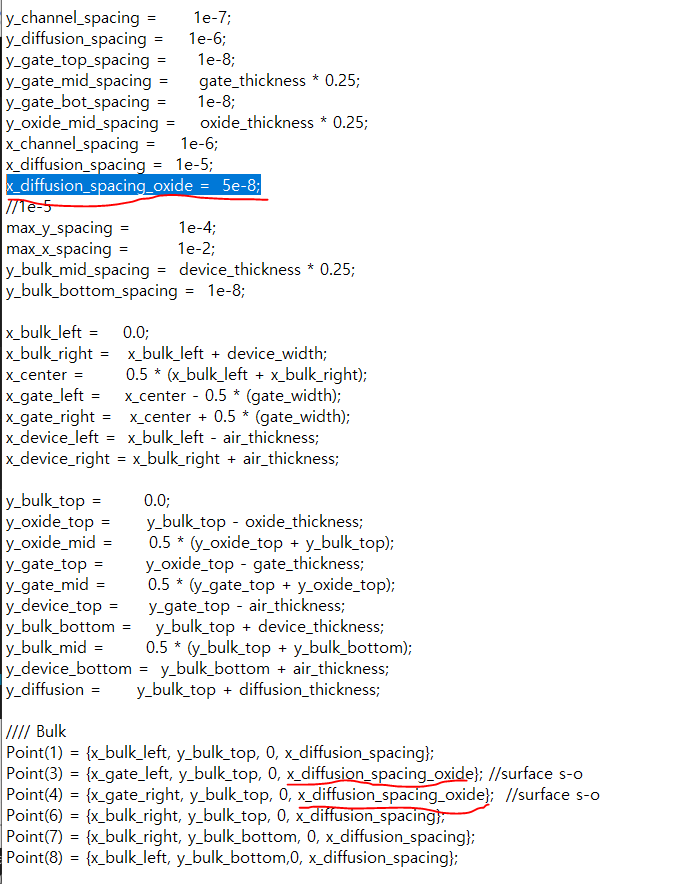
I am currently working toward providing the scripts used for publication.
For now, this python function can be used to sweep the bias and save the currents at each bias point:
https://github.com/devsim/devsim/blob/r1.6.1/python_packages/ramp.py
Please note this version is newer than the one in current 1.6.0 release, and should replace the one in the python_packages directory. The rampbias function will sweep the bias and reduce the bias step if there is a convergence issue.
For example, this function will sweep the drain bias from the current bias to vd using a step of vdestep.
rampbias(device, "drain", vd, vdstep, 0.01, 25, 1e-4, 1e30, printAllCurrents)
The last argument is a callback function which can be used to store the data to a file or print to the screen. For example:
def PrintCurrents(device, contact):
'''
print out contact currents
'''
# TODO add charge
contact_bias_name = GetContactBiasName(contact)
electron_current= ds.get_contact_current(device=device, contact=contact, equation=ece_name)
hole_current = ds.get_contact_current(device=device, contact=contact, equation=hce_name)
total_current = electron_current + hole_current
voltage = ds.get_parameter(device=device, name=GetContactBiasName(contact))
data = (voltage, electron_current, hole_current, total_current)
print("{0}\t{1}\t{2}\t{3}\t{4}".format(contact, *data))
return data
def printAllCurrents(device):
'''
Prints all contact currents on device
'''
for c in ds.get_contact_list(device=device):
PrintCurrents(device, c)
If you want to do multiple sweeps, you would need to backup and restore the solution variables at the beginning of each sweep.
def save_backup():
# we are now at the initial bias
biases = ('drain', 'gate', 'source', 'body')
backup = {'regions' : {},
'contacts' : {},
}
for i in oxide_regions:
backup['regions'][i] = {}
for j in ("Potential",):
backup['regions'][i][j] = ds.get_node_model_values(device=device, region=i, name=j)
for i in silicon_regions:
backup['regions'][i] = {}
for j in ("Potential", "Electrons", "Holes"):
backup['regions'][i][j] = ds.get_node_model_values(device=device, region=i, name=j)
backup['biases'] = {}
for i in biases:
backup['biases'][GetContactBiasName(i)] = ds.get_parameter(device=device, name=GetContactBiasName(i))
return backup
def restore_backup(backup):
for region, solution_data in backup['regions'].items():
for name, values in solution_data.items():
ds.set_node_values(device=device, region=region, name=name, values=values)
for contact_bias, value in backup['biases'].items():
print("RESTORE %s %g" % (contact_bias, value))
ds.set_parameter(device=device, name=contact_bias, value=value)
I hope to publish the scripts used for publication in the near future.